Very Short Answer Type Questions
Question. Give an example of a codon having dual function.
Answer : AUG.
Question. Name the negatively charged and positively charged components of a nucleosome.
Answer : Negatively charged component is DNA, positively charged component is histone octamer.
Question. Name one amino acid, which is coded by only one codon.
Answer : Methionine / Tryptophan
Question. Write the two specific codons that a translational unit of m-RNA is flanked by one on either sides.
Answer : Start codon – AUG Stop codon – UAA/UGA/UAG
Question. Write the dual purpose served by Deoxyribonucleoside triphosphates in polymerization.
Answer : Acts as a substrate, provide energy (from the terminal two phosphates).
Question. Retroviruses have no DNA. However the DNA of the infected host cell does possess viral DNA.
How is it possible ?
Answer : RNA is the genetic material in retrovirus. This RNA forms DNA by the process of reverse transcription with the help of the enzyme called reversetranscriptase.
Question. Name the enzyme and state its property that is responsible for continuous and discontinuous replication of the two strands of a DNA molecule.
Answer : DNA polymerase. Because it has exonuclease activity in 5’ → 3’ direction.
Question. Why is RNA more reactive in comparison to DNA?
Answer : 2′ – OH Present in RNA (in every nucleotide) make it reactive.
Question. Write the conclusion Griffith arrived at the end of his experiment with Streptococcus pneumoniae.
Answer : He concluded that the R-strain bacteria somehow been transformed by heat – killed S-strain bacteria, this must be due to transfer of genetic material
Question. Name the transcriptionally active region of chromatin in a nucleus.
Answer : Euchromatin i.e. lightly stained transcriptionally active region.
Question. Which of the two subunits of ribosome encounters an mRNA ?
Answer : The small subunit of ribosome encounters an mRNA.
Question. What is a cistron ?
Answer : Cistron is a segment of gene which codes for a certain polypeptide or protein.
Question. Name the specific components and the linkages between them that form deoxyguanosine.
Answer : Components : Deoxyribose sugar and nitrogen base guanine.
Linkage : N-glycosidic linkage.
Question. Name the enzyme involved in the continuous replication of DNA strand. Mention the polarity of the template strand.
Answer : DNA polymerase is involved in the continuous replication of DNA strand. Polarity of the strand is 3’ → 5’.
Question. How is repetitive/satellite DNA separated from bulk genomic DNA for various genetic experiments ?
Answer : Density gradient centrifugation.
Question. Why is it not possible for an alien DNA to become a part of a chromosome anywhere along its length and replicate normally ?
Answer : This DNA must be linked to ori/origin of replication site to start replication.
Question. Which one out of Rho factor and Sigma factor acts as the initiation factor during transcription in a prokaryote ?
Answer : Sigma factor (σ)
Question. Name the component a and b in the nucleotide with a purine given below.

Answer : a— Phosphate group
b— Nitrogenous base
Question. Mention one difference to distinguish an exon from an intron.
Answer : Exon : Coded/expressed sequence of nucleotides in mRNA.
Intron : Intervening sequence of nucleotides not appearing in processed mRNA.
Question. Name the enzyme that joins the small fragments of DNA of a lagging strand during DNA replication.
Answer : DNA ligase.
Question. Name the enzyme that transcribes hn-RNA in eukaryotes.
Answer : RNA Polymerase II.
Question. Which one is tailed with adenylate residues between 3’ and 5’ end of hnRNA ?
Answer : 3’ end is tailed with polyadenylate residues.
Question. A region of a coding DNA strand has the following nucleotide sequence : – ATGC – What shall be the nucleotide sequence in
(i) sister DNA segment it replicates, and
(ii) m-RNA polynucleotide it transcribes ?
Answer : (i) – TACG – (ii) – UACG-
Question. Name the specific components and the linkage between them that form deoxyadenosine.
Answer : Components : Nitrogen base Adenine (a purine) + Deoxyribose sugar.
Linkage : N-Glycosidic linkage [C – N – C].
Question. Name the types of synthesis ‘a’ and ‘b’ occurring in the replication fork of DNA as shown below :

Answer : a— Leading strand (continuous)
b—Lagging strand (discontinuous)
Question. Name the enzyme that helps to join DNA fragments.
Answer : DNA ligase joins/seal or stick the DNA fragments
Short Answer Type Questions – l
Question. Discuss the role the enzyme DNA ligase plays during DNA replication.
Answer : (Discontinuous) DNA fragments, are joined/sealed by them//sticky ends of vector and foreign DNA, joined by them. The following diagram can be considered in lieu of explanation.

Question. Why does hnRNA need to undergo splicing? Where does splicing occur in the cell ?
Answer : hnRNA has both exons and introns, Introns are non-coding regions, which are removed by the process called splicing, splicing occurs in the nucleus.
Question. How do histones acquire positive charge ?
Answer : Histones are rich in basic amino acids Lysine, Arginine (present as residues in their side chains), which are positively charged.
Question. State the functions of Ribozyme and release factor in protein synthesis respectively.
Answer : Ribozyme–helps in peptide bond formation.
Release factor–terminates translation/release polypeptide from ribosome.
Question. State the dual role of deoxyribonucleoside triphosphates during DNA replication.
Answer : The dual role of deoxyribonucleoside triphosphates are:
1. It serves are substrates for DNA synthesis
2. It provides energy for polymerisation reaction.
Question. Explain the two factors responsible for conferring stability to double helix structure of DNA.
Answer : The two factors responsible for conferring stability to double helix structure of DNA are:
1. Stacking of one base over another
2. H-bonding between the nitrogenous base
Question. Protein synthesis machinery revolves around RNA but in the course of evolution it was replaced by DNA. Justify A
Answer : Since RNA was unstable and prone to mutations, DNA evolved from RNA with chemical modifications that make it more stable. DNA has double stranded nature and has complementary strands. This further resist changes by evolving a process of repair.
Question. A template strand is given below. Write down the corresponding coding strand and the mRNA strand that can be formed along with their polarity.
3’ ATGCATGCATGCATGCATGCATGC 5’
Answer : Coding strand–
5′ TACGTACGTACGTACGTACGTACG 3’
mRNA strand–
5′ UACGUACGUACGUACGUACGUACG 3’
Question. Although a prokaryotic cell has no defined nucleus, yet DNA is not scattered throughout the cell. Explain.
Answer : DNA is negatively charged, positively charged protein, holds it in place, in large loops (in a region termed as nucleiod).
Question. Differentiate between a cistron and an exon.
Answer : Difference between cistron and an exon :
Cistron : A segment of DNA coding for a polypeptide chain, the structural gene in a transcription unit could be monocistronic (mostly in eukaryotes) or polycistronic (mostly in bacteria or prokaryotes).
Exon : The coding sequences or expressed sequences are defined as exons. Exons are said to be those sequences that appear in mature or processed mRNA.
Question. State the difference between the structural genes in a Transcription Unit of Prokaryotes and Eukaryotes.
Answer :

Question. Make a labelled diagram of an RNA dinucleotide showing its 3’ –5’ polarity.
Answer : Labelled diagram of RNA dinucleotide showing its 3′ – 5′ polarity is as follows :

Short Answer Type Questions – ll
Question. (i) Differentiate between a template strand and coding strand of DNA.
(ii) Name the source of energy for the replication of DNA.
Answer :

Question. Why is DNA molecule considered as a better hereditary material than RNA molecule ?
OR
Why is DNA a better genetic material when compared to RNA ?
Answer : DNA molecule is a better hereditary material as:
(i) It is more stable (due to presence of thymine and not uracil as in RNA).
(ii) Less reactive than RNA (as RNA has 2’ – OH making it more reactive).
(iii) Being less reactive , DNA is not easily degradable (RNA being more reactive is easily degradable).
(iv) Rate of mutation is slow (Rate of mutation in RNA is faster).
Question. (i) Why did Hershey and Chase use radioactive sulfur and radioactive phosphorus in their experiment ?
(ii) Write the conclusion they arrived at and how.
Answer : (i) Sulphur is a component of protein and thus would label the protein coat. Phosphorus is component of DNA.
(ii) Bacteria which were infected with viruses having radioactive DNA were found to contain radioactive DNA later on.
Bacteria which were infected with viruses having radioactive protein coat were not found to contain radioactivity. Conclusion-DNA is the genetic material.
Question. (i) Write what DNA replication refers to.
(ii) State the properties of DNA replication model.
(iii) List any three enzymes involved in the process along with their functions.
Answer : (i) DNA synthesis i.e. copying of DNA
(ii) (a) Semi-conservative.
(b) Semi-discontinuous.
(c) Unidirectional.
(iii) DNA polymerase III – adds nucleotides.
DNA polymerase I – fills the gaps.
RNA primase – brings primers.
Topoisomerase – causes unwinding.
DNA ligase – joins Okazaki fragments.
Question. (i) Given below is a single stranded DNA molecule. Frame and label its sense and antisense RNA molecule.
5’ ATGGGGCTC 3’ sense
(ii) How the RNA molecules made from above DNA strand help in silencing of the specific RNA molecules ?
Answer : (i) 5’ ATGGGGCTC 3’ sense
3’ TACCCCGAG 5’ antisense
RNA 5’ AUGGGGCUC 3’ sense
3’ UACCCCGAG 5’ antisense
(ii) The two strands of RNA (i.e. sense and antisense) being complementary will bind,with each other and form double stranded RNA. As a result its translation and protein,expression would be inhibited.
Question. Describe the experiments that established the identity of ‘transforming principles‘ of Griffith.
Answer : (i) Purification of biochemicals like Proteins, RNA & DNA from S cells (heat killed).
(ii) Presence of Protein & RNA in medium did not affect transformation.
(iii) DNA alone from S Bacteria caused R Bacteria to transform.
(iv) Digestion with DNAase inhibits transformation.
Conclusion : DNA is the transforming chemical / biochemical material as it was transferred from virus to bacteria.
Question. List the salient features of double helix structure of DNA.
Answer : DNA helix is made up of two polynucleotide chains, each constituted by sugar-phosphatebases, the chains are antiparallel in polarity (5’ → 3’ and 3’ → 5’), the bases are linked with H-bonds, Adenine pair with Thymine with two H-bonds while Guanine pair with Cytosine with three H-bonds, Coiling of the chain are in right handed fashion, pitch of the helix is 3.4 nm and there are 10 bp per turn, the plane of one base pair stack over the other in a double helix.

Question. Explain the role of DNA-dependent RNA polymerase in transcription.
Answer : RNA polymerase catalyzes RNA synthesis. To transcribe a gene, RNA polymerase proceeds through a series of well-defined steps, which are grouped into three phases : initiation, elongation and termination.
(i) Initiation : RNA polymerase binds with the promoter region of the DNA. The promoter – polymerase complex undergoes structural changes required for transcription.
(ii) Elongation : During elongation, RNA polymerase unwinds the DNA in front and re-anneals it behind, it dissociates the growing RNA chain from the template as it moves along. It also performs proof reading function.
(iii) Termination : Once the polymerase has transcribed the length of the gene, it stops and releases the RNA product.
Question. (i) Construct a complete transcription unit with promotor and terminator on the basis of the hypothetical template strand given below :

(ii) Write the RNA strand transcribed from the above transcription unit along with its polarity.
Answer :
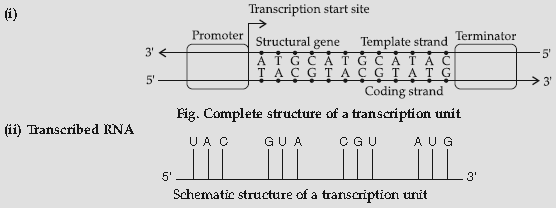
Question. (i) Name the enzyme that catalyses the transcription of hnRNA.
(ii) Why does the hnRNA need to undergo changes ?
List the changes hnRNA undergoes and where in the cell such changes take place.
Answer : (i) RNA polymerase II.
(ii) Has (non-functional) introns. (Methyl guanosine tri-phosphate is added to 5’ end) capping, tailing (Poly A tail at 3’ end added),
splicing (introns are removed and exons are joined). Nucleus.
Question. The base sequence in one of the strands of DNA is TAGCATGAT.
(i) Give the base sequence of its complementary strand.
(ii) How are these base pairs held together in a DNA molecule ?
(iii) Explain the base complementarity rules. Name the scientist who framed this rule.
Answer : (i) ATCGTACTA
(ii) Through Hydrogen bonds between A and T and C and G on the two strands.
(iii) A = T and C → G, Watson and Crick / Chargaff.
Question. (i) Why did Meselson and Stahl used 14N and 15N isotopes as the sources of nitrogen present in the culture medium in their experiment ? Explain.
(ii) Write the conclusion drawn by them from the experiment.
Answer : (i) Meselson and Stahl used 14N and 15N isotopes as the sources of nitrogen present in the culture medium in their experiment as nitrogen is a major constituent of DNA. Moreover 15N is by far the most abundant isotope of nitrogen and DNA with the heavier 15N isotope is also functional. E.coli can be grown for several generations in a medium with 15N easily. When DNA is extracted from these cells and centrifuged on a salt density gradient, the DNA separates out at the point at which its density equals that of the salt solution.
(ii) The experiment proves the semi-conservative nature of replication of DNA. In this type of replication, one strand of daughter DNA is new and one strand is old. It means, one strand of daughter duplex is derived from the old DNA, while the other strand is formed new. It proves semi-conservative replication of DNA.
Question. Identify A, B, C, D, E and F in the following table.

Answer : (I) A – Nitrogenous base / A – Pentose sugar.
B – Pentose Sugar / B – Nitrogenous base.
C – N-glycosidic linkage.
(II) D – phosphate group.
E – phospho ester linkage.
(III) F – (3′ – 5′) phosphodiester linkage.
Question. Name the specific enzyme responsible for nucleotide polymerisation in DNA replication. Write two characteristic features of this enzyme. Name the region on E. coli DNA where this enzyme can initiate replication.
Answer : (i) DNA – dependent DNA polymerase.
(ii) The enzyme uses DNA template to catalyse the polymerisation of deoxyribonucleotides.
(iii) Have to catalyse the reaction with high accuracy.
(iv) Origin of replication / Ori.
Question. Describe the termination process of transcription in bacteria.
Answer : (i) RNA polymerase binds to the promotor and initiates transcription. It uses nucleotide triphosphates as substrates and polymerises in a template dependent fashion following the rule of complementary. It somehow also facilitates opening of the helix structure and continues elongation. Only a short stretch of RNA remains bound to the enzyme. Once the polymerase reaches the terminator region, the nascent RNA falls off, also the RNA polymerase. This results into termination of the transcription.
(ii) RNA polymerase associates transiently with termination factor (rho) to terminate the transcription. Association of this factor alters the specificity of the RNA polymerase to terminate

Question. Name the three RNA polymerases found in eukaryotic cells and mention their functions.
Answer : RNA polymerase – I, transcribes rRNAs (28S -18S and 5.8S).
RNA polymerase – II, transcribes precursor of mRNA / hnRNA / heterogeneous RNA.
RNA polymerase – III, transcribes tRNA / 5Sr RNA / snRNA.
Question. (a) Mention two events in which DNA is unzipped.
(b) Predict the consequences when both the template and the coding strands of a DNA segment participate in transcription process?
Answer : (a) Replication and Transcription.
(b) If both the stands take part in transcription,
(i) One segment of DNA would be coding for two different proteins which will complicate the genetic information machinery
(ii) Two RNA molecules will be produced, complementary to each other, hence form a double stranded RNA, translation would not be possible.
Question. A DNA segment has a total of 1500 nucleotides out of which 410 are guanine containing nucleotides.
How many Pyrimidine bases this DNA segment possesses.
Answer : ∴ G = 410 ∴ C = 410 (because C = G)
∴ G + C = 410 + 410 = 820
∴ A + T = 1500 – 820 = 680
Question. (i) If the sequence of the coding strand in transcription unit is written as follows :
5’—ATGCATGCATGCATGCATGCATGCATGCAT
G—3’ write down the sequence of mRNA.
(ii) How is repetitive/satellite DNA separated from bulk genomic DNA for various genetic experiments.
(iii) Mention the polarity of DNA strands a-b & c-d shown in the replicating fork given below :

Answer : (i) 5’—AUGCAUGCAUGCAUGCAUGCAUGCAUG —3′
(ii) By density gradient centrifugation.
(iii) Polarity of DNA strand a–b is 3’→5′ & c–d has polarity 5’→3′.
Question. Explain the post transcriptional modifications the hn-RNA undergoes in eukaryotic cell.
Answer : • Splicing , Introns are removed and exons are joined.
• Capping, Methyl guanosine triphosphate / mGPPP is added to the 5’ end of hnRNA.
• Tailing, Polyadenylate residues are added to end in a template independent manner.

Thus the pyrimidine bases C + T
= 410 + 340
= 750
Question. Which one of these an intron and an exon is reminiscent of antiquity ?
Answer : The primary transcript in eukaryotes contain both exons & introns (the non coding sequences). The introns are removed during processing by splicing & the exons are joined. The presence of introns in primary transcript is the reminiscent of antiquity
Question. Name two basic amino acids that provide positive charges to histone proteins.
Answer : Histone proteins have a positively charged surface because of the presence and abundance of two basic amino acids : (i) Lysine and (ii) Arginine as compared to other amino acids.
Question. If the base adenine constitute 30% of an isolated DNA fragment, then what is the expected percentage of the base cytosine in it.
Answer : As A = 30%
T = 30% (as A = T)
A + T = 60%
G + C = 100 – 60 = 40%
∴ C (Cytosine) =20% (as C = G)
Long Answer Type Questions
Question. (i) Describe the series of experiments of F. Griffith. Comment on the significance of the results obtained.
(ii) State the contribution of Macleod, McCarty and Avery.
OR
(i) Explain with the help of Griffith’s experiment how the search for genetic material was conducted and what was the conclusion drawn ?
(ii) How did Macleod, McCarty and Avery establish the biochemical nature of the so called “genetic material” identified by Griffith in his experiment.
OR
(i) Describe the various steps of Griffith’s experiment that led to the conclusion of the ‘Transforming principle’.
(ii) How did the chemical nature of the ‘Transforming principle’ get established ?
OR
Describe Frederick Griffith’s experiment on Streptococcus pneumoniae. Discuss the conclusion he arrived at.
Answer : (i) Streptococcus pneumoniae :
S – Strain (virulent) injected into mice → mice die.
R – strain injected into mice → mice alive.
S – strain (heat killed) injected into mice → mice alive.
R – strain (alive) + S (heat killed) strain inject into mice → mice die.
R strain (non-virulent) picked up genetic material from S strain (virulent) and get transformed.
(ii) They (worked on the bio-chemical nature of transforming principle in Griffith’s experiment) purified proteins, DNA and RNA from heat killed S cells, they discovered protein digesting enzyme (protease), RNA digesting enzyme (RNAase) did not affect transformation, Digestion with DNase inhibited transformation, concluded DNA is the heredity material.
Question. Answer the following questions based on Hershey and Chase experiments :
(i) Name the kind of virus they worked with and why ?
(ii) Why did they use two types of culture media to grow viruses in ? Explain.
(iii) What was the need for using a blender and later a centrifuge during their experiments ?
(iv) State the conclusion drawn by them after the experiments. U [Delhi Set-III, 2016]
Answer : (i) Bacteriophage, they infect bacteria.
(ii) Two types of culture media were used in order to make protein of viruses (with the help of S35) radioactive in one case, and DNA molecule in virus (with the help of P32 radioactive in other case. so as to identify which one of the two had entered into the bacteria during viral infection.
(iii) Blender : To separate the viral protein coats that are still attached to the surface of bacteria.
Centrifuge : To separate lighter supernatant (containing viral protein coats) from denser residue (containing bacteria). ½
(iv) DNA is the genetic material i.e. passed from virus to bacteria.
Question. Describe the packaging of DNA helix in a prokaryotic cell and an eukaryotic nucleus.
Answer : Prokaryotes : Negatively charged DNA is held with positively charged proteins in nucleoid, DNA in nucleoid is organised in large loops held by protein.
Eukaryotes : In nucleus, the negatively charged
DNA is wrapped around positively charged histone octomer to form nucleosome, nucleosomes are repeated to constitute chromatin at higher level, additional set of non-histone chromosomal protein gets associated with chromatin.
Question.

(i) Identify strands ‘A’ and ‘B’ in the diagram of transcription unit given above and write the basis on which you identified them.
(ii) State the functions of Sigma factor and Rho factor in the transcription process in a bacterium.
(iii) Write the functions of RNA polymerase-I and RNA polymerase-III in eukaryotes.
Answer : (i) A-Template strand
B-Coding strand
Template strand has polarity 3’-5’
Coding strand has polarity 5’-3’
On the basis of polarity with respect to promoter.
(ii) Sigma factor associates with RNA polymerase to initiate transcription, Rho factor gets associated to RNA polymerase to terminate transcription.
(iii) RNA polymerase I – Transcribes -RNAs ½ RNA polymerase III – Transcribes tRNA / 5srRNA / 5nRNA
Question. How does the flow of information in HIV deviate from the Central Dogma proposed by Francis Crick.
Answer : According to Central Dogma of molecular biology, there is unidirectional flow of genetic information from DNA to m RNA and from here to protein.

But in HIV (Human immuno-deficiency virus) there is central dogma reverse i.e. the flow of genetic information is in reverse direction. It is because the virus contains the genetic RNA and produces an enzyme reverse transcriptase. This enzyme helps to synthesize DNA from genetic RNA of HIV by a process called as reverse transcription or Teminism. This newly synthesized DNA functions as master copy producing mRNA through transcription & RNAs controlling translation to synthesize protein.

The phenomenon of reverse transcription was discovered by Temin & Baltimore (1970) in Retrovirus
Question. (i) Hershey and Chase carried their experiment in three steps : infection, blending, centrifugation.
Explain each step.
(ii) Write the conclusion and interpretation of the result they obtained.
OR
Describe the Hershey and Chase experiment.
Write the conclusion drawn by the scientists after their experiment.
OR
How did Alfred Hershey and Martha Chase arrive at the conclusion that DNA is the genetic material ?
Answer : (i) Infection : Radioactive phosphorus / phosphorus labelled bacteriophages were allowed to infect E.coli – growing in a culture medium, simultaneously radioactive sulphur / Sulphur labelled bacteriophage was allowed to infect E.coli growing in another culture medium.
(a) Blending : As infection proceeds – the viral coats are removed from the bacteria by agitating in a blender
(b) Centrifugation : virus particles were separated from bacteria by spinning them in a centrifuge.
(ii) Conclusion : DNA is the genetic material. Interpretation : Sulphur labelled viral protein did not enter the bacteria during infection, whereas phosphorus labelled viral DNA entered into the bacteria to cause infection.
Question. (i) How are the following formed and involved in DNA packaging in a nucleus of a cell ?
(a) Histone Octamer,
(b) Nucleosome,
(c) Chromatin.
(ii) Differentiate between Euchromatin and Heterochromatin.
Answer : (i) (a) Eight molecules of positively charged basic proteins called histones are organised to form histone octamer.
(b) Negatively charged DNA wrapped around positively charged histone octamer to give rise to nucleosome.
(c) Nucleosome constitute the repeating unit of a structure called chromatin.

Question. (i) Describe the process of transcription in bacteria.
(ii) Explain the processing the hnRNA needs to undergo before becoming functional mRNA in eukaryotes.
OR
Explain the process of transcription in a prokaryote.
Answer : (i) Bacteria transcription is the process in which messenger RNA transcripts of genetic material in bacteria are produced to be translated for the production of proteins. Bacterial transcription occurs in the cytoplasm alongside translation. The process of transcription is completed in three steps : Initiation, elongation and termination.
(a) Initiation : The enzyme binds at the promoter site of DNA and initiates the process of transcription. It causes the local unwinding of the DNA double helix. An initiation sigma factor (σ) present in RNA polymerase initiates the RNA synthesis.
(b) Elongation : The RNA chain is synthesized in the 5’-3’ direction. RNA polymerase uses nucleoside triphosphate as substrate and polymerisation occurs according to complementarity.
(c) Termination : Termination occurs when termination factor (rho) alters the specificity of RNA polymerase to terminate the transcription.
As the RNA polymerase proceeds to perform elongation, a short stretch of RNA remains bound to the enzyme. As the enzyme reaches the termination region, this nascent RNA falls off and transcription is terminated.
(ii) The precursor of mRNA i.e. hnRNA contains both introns and exons. Introns are removed and exons are joined by a process called splicing. The remaining mRNA is processed in two ways :
(a) Capping : Here, an unusual nucleotide called methyl guanosine triphosphate (cap) is added to the 5’ end of hnRNA.
(b) Tailing : Here, adenylate residues (200-300) are added at 3’ end of hnRNA in a template independent manner.
When hnRNA is fully processed, it is known as mRNA, which is transported out of the nucleus to get translated.
Question. (a) Why does DNA replication occur in small replication forks and not in its entire length ?
(b) Why is DNA replication continuous and discontinuous in a replication fork ?
(c) State the importance of origin of replication in a replication fork.
Answer : (a) DNA being very long , requires high energy for opening along its entire length.
(b) DNA dependent DNA polymerase catalyse polymerisation only in one direction i.e. 5’ 3’.
Two strands of DNA are anti parallel and have opposite polarity.
(c) Site where replication originates
Question. (a) State the ‘Central dogma’ as proposed by Francis Crick. Are there any exceptions to it ? Support your answer with a reason and an example.
(b) Explain how the biochemical characterisation (nature) of “Transforming Principle” was determined, which was not defined from Griffith’s experiments.
Answer : (b) Protein, DNA and RNA were purified from heat killed S strain / smooth Streptococcus pneumonia = ½
Protein + Protease → transformation occurred
(R cell to S type) = ½
RNA + RNA ase → transformation occurred
(R cell to S type) = ½
DNA + DNA ase → transformation inhibited = ½
Hence DNA alone is the transforming material = ½
TOPIC-2
Genetic Code, Translation, Lac Operon, Human
Genome Project and DNA Fingerprinting
Very Short Answer Type Questions
Question. Mention the contribution of genetic maps in human genome project.
Answer : Sequencing of genes, DNA finger printing, tracing human history, chromosomal location for disease associated sequences. (Any one)
Question. Mention the role of the codons AUG and UGA during protein synthesis.
Answer : AUG – codes for methionine / initiation codon.
UGA – termination codon / stop codon.
Question. Mention two applications of DNA-polymorphism.
Answer : Genetic mapping & DNA-finger printing.
Question. State which human chromosome has
(i) The maximum number of genes and
(ii) The one which has the least number of genes.
Answer : (i) Chromosome no. 1 : 2968 genes
(ii) Y chromosomes : 231 genes
Short Answer Type Questions – l
Question. What is aminoacylation ? State its significance.
Answer : Amino acids are activated in the presence of ATP and linked to (cognate) t-RNA.
Carries amino acid to the site of synthesis/reaches amino acids to the respective codon.
Question. State the roles of AUG codon at 5′ end and UAG at 3′ end of a certain m-RNA during translation.
Answer : AUG codon at 5′ end = Start codon (for translation)/codes for methionine.
UAG codon at 3′ end = Stop codon (for translation)/ terminate polypeptide chain.
Question. Following are the features of genetic codes. What does each one indicate ? Stop codon, Unambiguous codon, Degenerate codon, Universal codon.
Answer : The features of genetic code are :
(i) Stop codon : does not code for any amino acid / terminates the synthesis of polypeptide chain.
(ii) Unambiguous codon : one codon codes for one amino acid only.
(iii) Degenerate codon : some amino acid are coded by more than one codon.
(iv) Universal codon : genetic code is same for all organisms (bacteria to humans).
Question. (i) Name the scientist who suggested that the genetic code should be made of a combination of three nucleotides.
(ii) Explain the basis on which he arrived at this conclusion.
Answer : (i) George Gamow.
(ii) There are four bases and 20 amino acids.
(There should be atleast 20 different genetic codes for these 20 amino acids). Only possible combinations that would meet the requirement in combinations of 3 bases that will give 64 codons.
Question. One of the salient features of the genetic code is that it is nearly universal from bacteria to humans.
Mention two exceptions to this rule. Why are some codons said to be degenerate ?
Answer : (i) Mitochondrial codons.
(ii) Some protozoans. Since some amino acids are coded by more than one codon hence it is called as degenerate.
Question. Write the full form of VNTR. How is VNTR different from ‘Probe’ ?
Answer : VNTR – Variable Number Tandem Repeats. Probe – is labelled / radio active (single stranded hybridised DNA fragments).
Question. Where does peptide bond formation occur in a bacterial ribosome and how ?
Answer : Between the two amino acids (found on charged tRNA), bound to the two sites of the large sub units of bacterial ribosomes, when two charged tRNAs are brought close enough, peptide bond is formed with the help of ribozyme.
Question. ‘Degenerate‘ and ‘Universal‘ are salient features of a genetic code. Explain.
Answer : Degenerate : Same amino acids are coded by more than one codon.
Universal : One codon shall code for the same amino acid in all organisms (UUU would code for phenylalanine from bacteria to human beings).
Question. Why does the lac operon shut down some time after the addition of lactose in the medium where E.coli was growing? Why low level expression of lac operon is always required?
Answer : After addition of lactose, complete breakdown of lactose to glucose and galactose takes place.
Therefore, there is no more lactose to bind to the repressor protein and the lac operon shuts down.
A very low level of expression of lac operon has to be present in the cell all the time, otherwise lactose cannot enter the cells.
Short Answer Type Questions – ll
Question. Write any three goals of Human Genome Project.
Answer : The three main goals of HGP are :
(i) To determine the sequences of 3 billion base pairs that make up the human DNA.
(ii) To identify all the estimated genes in human DNA.
(iii) To store this information in databases.
Question. Where is an ‘operator’ located in a prokaryote DNA ? How does an operator regulate gene expression at transcriptional level in a prokaryote ? Explain.
Answer : The operator region is located adjacent to promoter elements / prior to structural gene.
In regulation of gene expression :
switch off – the repressor binds to the operator region and prevents transcription.
switch on – In the presence of inducer the repressor is inactivated (by the interaction with the inducer), operator allows RNA polymerase access to the promoter and transcription proceeds.
Question. Following the collision of two trains a large number of passengers are killed. A majority of them are beyond recognition. Authorities want to hand over the dead to their relatives. Name a modern scientific method and write the procedure that would help in the identification of kinship.
Answer : DNA fingerprinting is used for identification of kinship. Procedure :
(i) Variable number of tandem repeats (VNTR’s) are satellite DNA’s that show high degree of polymorphism. They are used as probes in DNA
fingerprinting.
(ii) Fragments of DNA from an individual are isolated and cut with restriction endonucleases.
(iii) Fragments are separated according to their size and molecular weight through gel electrophoresis.
(iv) Fragments separated through electrophoresis gel are blotted (immobilised) on a synthetic membrane such as nylon or nitrocellulose.
(v) Immobilised fragments are hybridised with a VNTR probe.
(vi) Hybridised DNA fragments can be detected by autoradiography.
(vii) VNTRs are different in size, ranging from 0.1 to 20 kb. Hence, in the autoradiogram, a band of different sizes will be obtained.
(viii) These bands are the characteristic feature of an individual. They are different in each and every individual except identical twins.
Question. (i) How many codons code for amino acids and how many are unable to do so ?
(ii) Why are codes said to be (i) degenerate and (ii) unambiguous ?
Answer : (i) Sixty one, Three.
(ii) (a) Degenerate—One amino acid may be coded by several codons.
(b) Unambiguous or specific—Each codon codes for a specific amino acid.
Question. How are the structural genes activated in the lac operon in E. coli ?
Answer : Lactose acts as the inducer, binds with repressor protein, frees operator gene, RNA polymerase freely moves over the structural genes, transcribing lac mRNA, which in turn produces the enzymes responsible for the digestion of lactose.
A complete labelled diagram depicting the concept can be evaluated in lieu of explanation.
Question. (i) What do ‘Y’ and ‘B’ stand for in ‘YAC’ and ‘BAC’ used in Human Genome Project (HGP). Mention their role in the project.
(ii) Write the percentage of the total human genome that codes for proteins and the percentage of discovered genes whose functions are known as observed during HGP.
(iii) Expand ‘SNPs’ identified by scientists in HGP.
Answer : (i) Y = Yeast
B = Bacterial sed as vector for cloning foreign DNA ½
(ii) (<) 2% , (<) 50%.
(iii) Single Nucleotide Polymorphism
Question. Given below are the sequence of nucleotides in a particular mRNA and amino acids coded by it : UUU AUG UUC GAG UUA GUG UAA
Phe –Met –Phe –Glu –Leu –Val Write the properties of the genetic code that can be and that cannot be correlated from the above given data.
Answer : Properties of genetic code that can be correlated are:
(i) The codon is a triplet e.g. UUU, AUG etc. They form triplets.
(ii) One codon codes for only one amino acid and not other hence it is unambiguous and specific.
(iii) AUG has a dual function as it codes for methionine and also acts as the initiator codon.
(iv) UAA is the stop codon. It codes for valine.
(v) Code is commaless, continuous and does not have pauses.
(vi) The sequence of triple N-bases in mRNA corresponds to the sequence of amino-acids in a polypeptide.
Property that cannot be correlated is that mostly AUG work as an initiating codon.
Question. Unambiguous, universal and degenerate are some of the terms used for the genetic code. Explain the salient features of each one of them.
Answer : Unambiguous – One codon codes for one amino
acid = e.g. AUG (methionine).
Universal – Codon and its corresponding amino acid are the same in all organisms.
Example : Bacteria to human UUU codes for phenylalanine (phe).
Degenerate – Some amino acids are coded by more than one codon. ½
Example : UUU and UUC code for phenylalanine(phe).
Question. (i) List the structural genes involved in the digestion of lactose in E.coli. Highlight the function of any one.
(ii) What triggers the transcription of these genes ?
Answer : (i) There are three structural genes (z, y, a) which transcribes a polycistronic mRNA.
(a) Gene z codes β-galactosidase (β-gal), which catalyses the hydrolysis of lactose into galactose and glucose.
(b) Gene y codes for permease, which increases the permeability of the cell to β-galactosidase (lactose).
(c) Gene a codes for transacetylase, which catalyses the transacetylation of lactose into its activeform.
(ii) RNA polymerase triggers the transcription of these genes.
Question. Differentiate between the following :
(i) Promoter and terminator in a transcription unit.
(ii) Exon and intron in an unprocessed eukaryotic mRNA.
(iii) Inducer and repressor in lac operon.
Answer : (i) Promotor and terminator in a transcription
unit : Promoter is located towards 5’ end while terminator is located towards 3’ end.
(ii) Exon and intron in an unprocessed eukaryotic mRNA :
Exon : The coding sequences or expressed sequences are defined as exons. Exons are said to be those sequences that appear in mature or processed RNA.
Introns : The exons are interrupted by introns. Introns or intervening sequences that do not appear in mature or processed RNA.
(iii) Inducer and repressor in lac operon :
Inducer : Lactose is the substrate for the enzyme beta-galactosidase and it regulates switching on and off of the operon. It is termed as Inducer.
Repressor : The repressor of the operon is synthesized (all the time constitutively) from the
gene. The repressor protein binds to the operator region of the operon and prevents RNA polymerase from transcribing the operon.
Question. (a) Explain VNTR and describe its role in DNA fingerprinting.
(b) List any two applications of DNA fingerprinting technique.
Answer : (a) VNTR-(i) Variable Number of Tandem Repeats.
(ii) used as a probe (because of its high degree of polymorphism).
(b) Forensic science / criminal investigation (any point related to forensic science) / determine population and genetic diversities / paternity testing / maternity testing / study of evolutionary biology.
Question. A criminal blew himself up in a local market when he was chased by cops. His face was beyond recognition. Suggest and describe a modern technique that can help establish his identity.
OR
A number of passengers were severely burnt beyond recognition during a train accident. Name and describe a modern technique that can help establish their identity.
OR
During a fire in an auditorium a large number of assembled guests got burnt beyond recognition. Suggest and describe a modern technique that can
help hand over the dead to their relatives.
Answer : DNA finger printing Isolation of DNA and digestion of DNA by restriction endonucleases, separation of DNA fragments by gel electrophoresis and transferring (blotting) of separated DNA fragments to synthetic membrane or nitrocellulose or nylon, hybridization using VNTR probe and detection of hybridised DNA fragments by autoradiography, matching the banding pattern so obtained with that of relative.
Long Answer Type Questions
Question. Explain sequentially the process of ‘Translation’ in a prokaryote. Name the cellular factory where this process occurs.
Answer : (i) Small subunit of ribosome encounters mRNA and protein synthesis process begins. There are two sites in the large subunit for subsequent amino acids to bind to and thus be close enough to each other for the formation of the peptide bond.
(ii) The order and sequence of the amino acid are defined by the bases in m-RNA.
(iii) In the first phase the amino acid are activated in the presence of ATP and linked to t-RNA(aminoacylation of t-RNA).
(iv) Ribozyme/Ribosome acts as a catalyst for formation of peptide bond.
(v) A translational unit has a sequence of RNA that is flanked by start codon (AUG) at 5′ end and stop codon at 3′ end (UAA, UAG, UGA).
(vi) Initiation–ribosome binds to m-RNA at start codon (recognized by tRNA).
(vii) Elongation phase: Amino acids linked to tRNA bind to appropriate codon of mRNA by complementary base pairing.
(viii) Ribosome moves codon to codon along mRNA, polypeptide sequence formed as dictated by DNA and represented by mRNA
(ix) At the end, termination occurs by a release factor which binds to stop codon releasing the polypeptide.
Ribosome is the cellular factory.
Question. (i) Write any two different levels at which regulation of Gene Expression could be exerted in Eukaryotes.
(ii) Give a labelled schematic representation of ‘‘lac operon’’ in its ‘‘Switched Off’’ position.
Answer : (i) Transcriptional level (formation of primary transcript).
• Processing level (regulation of splicing).
• Transport of mRNA from nucleus to cytoplasm.
• Translational level.

Question. (i) How is DNA fingerprinting done ? Name any two types of human samples which can be used for DNA fingerprinting. Explain the process sequentially.
(ii) Mention any two situations when the technique is useful.
Answer : (i) High degree of polymorphism forms the basis of DNA fingerprinting. It involves isolation of DNA, digestion of DNA by restriction endonucleases, separation of DNA fragments by electrophoresis, transferring/blotting of separated DNA fragments to synthetic membranes (nylon or nitrocellulose),
Hybridisation using labelled VNTR probes, detection of hybridised DNA fragments by autoradiography.
DNA from blood/hair follicle/skin/bone/saliva/ sperm.
(ii) Helps as identification tool in forensic applications, in determining population and genetic diversity and helps in paternity testing.
Question. Explain the process of protein synthesis from processed m-RNA.
Answer : For initiation, the ribosome binds to the mature m-RNA at the start codon (AUG) that is recongnized by the initiator t-RNA. During elongation, charged t-RNA sequentially binds to the appropriate codon in m-RNA with the aniticodon present on tRNA.
The ribosome moves from one codon to another adding amino acids one after the other to form polypeptide i.e. translation. During termination, the release factor binds to stop codon (UAA, UAG, UGA), terminating translation and releasing the polypeptide chain.
Question. (i) Explain the role of regulatory gene, operator, promoter and structural genes in lac operon when E. coli is growing in a culture medium with the source of energy as lactose.
(ii) Mention what would happen if lactose is withdrawn from the culture medium.
Answer : (i) Regulatory gene : codes for repressor of lac operon.
Operator : Provides site for binding of repressor
protein to prevent transcription.
Promoter : Provides site for binding of RNA polymerase.
Structural Genes : codes for enzymes/gene products required for metabolism of lactose.
(ii) If lactose is withdrawn from the culture medium the operon is not induced or expressed.
Question. Which methodology is used while sequencing the total DNA from a cell ? Explain it in detail.
Answer : Methodology used : Sequence Annotation :
(i) total DNA from a cell is isolated.
(ii) converted into random fragments of relatively smaller sizes
(iii) and cloned in suitable host using specialized vectors.
(iv) The cloning results in amplification of each piece of DNA fragment.
(v) The fragments are sequenced using automated DNA sequencers,
(vi) these sequences are then arranged based on some overlapping regions present in them.
(vii) This requires generation of overlapping fragments for sequencing.
(viii) Specialized computer based programmes are developed and
(ix) these sequences are subsequently annotated and assigned to each chromosome.
Question. (i) Describe the structure and function of a t-RNA molecule. Why is it referred to as an adapter molecule ?
(ii) Explain the process of splicing of hn-RNA in a eukaryotic cell.
Answer : (i) Clover-leaf shaped or inverted L shaped molecules has an anti codon loop with bases complementary to specific codon, has an amino acid acceptor end.
As it reads the code on one hand binds with the specific amino acid on the other hand.
(ii) Introns are removed, exons are joined in a definite order.
OR
Process of splicing shown diagrammatically.
Question. Name the major types of RNAs and explain their role in the process of protein synthesis in prokaryote
Answer : Three types of RNAs : (i) mRNA (ii) tRNA (iii) rRNA.
Role :
mRNA – Provides the template for protein synthesis by bringing the genetic information from
DNA to the site of protein synthesis / ribosome, also provides site to initiate and terminate the process of protein synthesis.
tRNA – Its anti codon loop read the genetic code on mRNA, brings the corresponding amino acid and bound its amino acid binding end on to the mRNA.
rRNA – Forms a structural component of ribosome, (23SRNA) acts as a catalyst / ribozyme for the formation of peptide bond.
Question. Write the different components of a lac-operon in E. coli., Explain its expression while in an ‘open‘ state.
OR
Explain the role of lactose as an inducer in a lac operon.
Answer : The arrangement where a (Polycistronic) structural gene is regulated by a common promoter and regulatory genes.
Question. (i) Name the scientist who postulated the presence of an adapter molecule that can assist in protein synthesis.
(ii) Describe its structure with the help of a diagram. Mention its role in protein synthesis.
Answer : (i) Francis Crick
(ii) Clover leaf / inverted L, Anticodon loop (complementary to codon of mRNA), acceptor end to bind amino acid. It reads the codons on mRNA with the help of anticodon loop, brings the corresponding amino acid for the formation of polypeptide chain.

Question. Describe the interaction of t-RNA, m-RNA and ribosomes during the events of translation.
Answer : (i) For initiation, the ribosome binds to the mRNA at the start codon / AUG.
(ii) Charged tRNA binds to the appropriate codon on mRNA forming complementary base pairs on tRNA as anti codon in the ribosome.
(iii) Ribosomes moves from codon to codon along mRNA, aminoacids are added one by one brought by tRNA, form the polypeptide chain.
